Predefined Antimicrobial Clinical Pathways and Formulary Rules
for Infectious Disease Lab Test Reports (Pathogen + AMR Detection Only)
Last updated: February 9, 2026
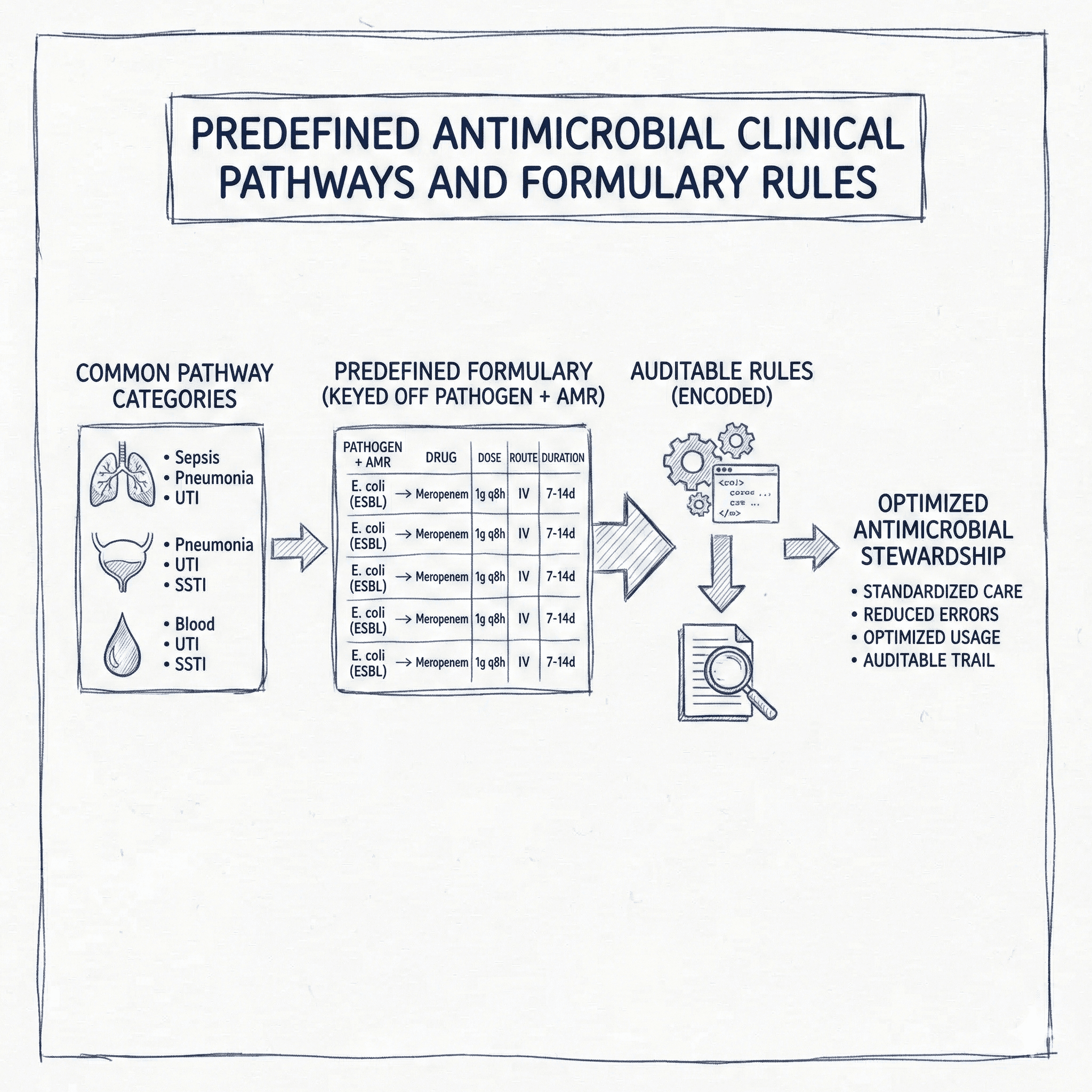
If you are generating an infectious disease test report that only knows two things—(1) the detected pathogen(s) and (2) detected antimicrobial resistance (AMR) markers—the safest and most operational way to provide treatment suggestions is to rely on:
- Predefined clinical pathways (organization-approved algorithms)
- Predefined formulary rules (machine-readable “if pathogen + AMR, then regimen(s)” entries)
This post summarizes common pathway categories used in real-world antimicrobial stewardship, then shows how a predefined formulary can be encoded as auditable rules (drug, dose, route, duration) keyed off pathogen + AMR.
What “Predefined” means in this context
- Predefined clinical pathways: Organization-approved algorithms (often syndrome- and setting-based) that tell clinicians what to start, when to narrow, and how long to treat.
- Predefined formulary rules: A curated library of “if pathogen + AMR pattern, then recommended regimen(s)” entries, typically used for targeted therapy once microbiology/AMR data is available.
- Guardrails are mandatory: local formulary availability, local antibiogram thresholds, renal/hepatic dosing adjustments, allergy rules, and an escalation path (e.g., infectious disease consult).
Common categories of predefined antimicrobial clinical pathways
Below are pathway categories stewardship programs commonly maintain. For each category, a sample pathway type is shown (1–2 examples).
1) Formulary & stewardship control
- Restricted/approval antibiotics list (e.g., carbapenems, linezolid, daptomycin require ID or pharmacy approval).
- Tiered antibiotic lists aligned to WHO AWaRe (Access/Watch/Reserve) to steer first-line choices.
2) Empiric therapy (syndrome-based)
- Community-acquired pneumonia (CAP) pathway (non-ICU vs ICU; risk factors for MRSA/Pseudomonas).
- Complicated UTI/pyelonephritis pathway (community vs healthcare-associated; sepsis flags).
3) Targeted therapy (pathogen + susceptibility / AMR-based)
- ESBL-producing Enterobacterales pathway (carbapenem vs beta-lactam/beta-lactamase inhibitors when appropriate).
- MRSA-confirmed pathway (vancomycin vs alternatives; bacteremia vs SSTI).
4) Setting-based care pathways
- Outpatient pathways (e.g., uncomplicated cystitis with oral-only regimens).
- Inpatient/ICU pathways (e.g., sepsis bundles: cultures first, broad coverage quickly, then narrow).
5) Prophylaxis pathways
- Surgical prophylaxis pathways (agent selection and timing relative to incision).
- Procedure/device prophylaxis pathways (e.g., select dental prophylaxis scenarios).
6) Duration, IV-to-PO switch, and de-escalation
- 48–72 hour antibiotic “time-out” pathway to reassess need and narrow therapy.
- IV-to-oral switch criteria pathways (hemodynamically stable, improving, tolerating PO).
7) Special populations
- Renal/hepatic dosing pathways (dose/interval by creatinine clearance; dialysis considerations).
- Pregnancy/pediatrics pathways (safe alternatives and age-specific dosing).
8) Infection control and AMR containment
- MDRO detection pathway (flag, isolation precautions triggers, stewardship review).
- Rapid diagnostic stewardship pathway (e.g., MALDI-TOF/BCID results trigger same-day antibiotic review).
9) Service-line or unit-specific pathways
- Oncology febrile neutropenia pathways (broad anti-pseudomonal beta-lactam first).
- Transplant prophylaxis/treatment pathways (e.g., CMV prophylaxis protocols).
10) Digital order sets and decision support
- EHR order sets that pre-populate antibiotic, dose, labs, and reassessment reminders.
- Stewardship dashboards that combine antibiogram + restricted antibiotic policy + alerting (e.g., prolonged therapy).
Predefined formulary rules keyed off pathogen + AMR
For a lab-driven report, “predefined formulary” becomes a rules library. Each rule is triggered by pathogen + AMR signature, and returns one or more candidate regimens with dose/route/duration, plus constraints.
Important: Site of infection and severity still matter. In practice, store multiple variants of the same rule by infection site (e.g., UTI vs bacteremia vs pneumonia), and include a “requires clinical context” warning when site is unknown.
Sample formulary entries (machine-readable clinical suggestions)
These examples illustrate what a single, predefined formulary entry can look like when the report detects only pathogen + AMR markers.
Example A: ESBL-producing E. coli (blaCTX-M) – Complicated UTI / Pyelonephritis
| Attribute | Definition |
|---|---|
| Trigger | Escherichia coli detected; ESBL marker blaCTX-M positive; carbapenemase markers (KPC/NDM/OXA-48-like) not detected |
| Clinical focus | Complicated UTI or pyelonephritis (targeted therapy after microbiology confirmation) |
| Preferred regimen | Ertapenem 1 g IV q24h × 7 days |
| Alternative regimen | Meropenem 1 g IV q8h × 7 days (use when broader coverage or severe illness is present) |
| Oral step-down option | If susceptible and clinically stable: TMP-SMX DS (160/800 mg) PO q12h to complete 10 days total therapy |
| Renal adjustment | If CrCl < 30 mL/min: ertapenem 500 mg IV q24h |
| Stewardship notes | Avoid ceftriaxone and piperacillin-tazobactam as definitive therapy when ESBL is confirmed; reassess at 48–72h and narrow when possible |
| Source examples | IDSA guidance for resistant Gram-negative infections; Sanford Guide antimicrobial therapy tables (latest digital edition) |
Example B: MRSA-confirmed (mecA) – Skin and soft tissue infection
| Attribute | Definition |
|---|---|
| Trigger | Staphylococcus aureus detected; mecA (or mecC) positive |
| Clinical focus | Cellulitis/abscess when MRSA is confirmed by molecular markers and consistent with clinical picture |
| Preferred regimen (IV) | Vancomycin 15 mg/kg IV q12h; target AUC 400–600 mg·h/L (or trough-based per local protocol). Duration: 5–10 days depending on response |
| Alternative regimen (PO, mild) | Doxycycline 100 mg PO q12h × 7–10 days (if appropriate and susceptible) |
| Alternative regimen (PO) | Clindamycin 300–450 mg PO q6–8h × 7–10 days if D-test negative and local susceptibility supports use |
| Stewardship notes | If mecA is negative and MSSA supported by culture, de-escalate to a beta-lactam |
| Source examples | IDSA SSTI guidance; Sanford Guide agent dosing tables |
Example C: CRE Enterobacterales with blaKPC – Bloodstream infection (high-risk)
| Attribute | Definition |
|---|---|
| Trigger | Enterobacterales detected (e.g., Klebsiella pneumoniae); carbapenemase blaKPC detected |
| Clinical focus | Suspected/confirmed bloodstream infection / severe infection where KPC is detected |
| Preferred regimen | Ceftazidime-avibactam 2.5 g IV q8h (2-hour infusion) × 10–14 days (guided by source control/response) |
| Alternative regimen | Meropenem-vaborbactam 4 g IV q8h (3-hour infusion) × 10–14 days (if available/appropriate) |
| Renal adjustment | Adjust per product labeling and pharmacy protocol; high variability across agents |
| Stewardship notes | Strongly consider ID consult. Ensure source control and repeat blood cultures per local policy |
| Source examples | IDSA guidance for CRE/KPC; FDA labeling for dosing and renal adjustment details |
Example D: Pseudomonas aeruginosa with metallo-beta-lactamase (e.g., blaVIM/blaNDM) – HAP/VAP
| Attribute | Definition |
|---|---|
| Trigger | Pseudomonas aeruginosa detected; metallo-beta-lactamase marker (blaVIM or blaNDM) detected |
| Clinical focus | Hospital-acquired or ventilator-associated pneumonia (requires clinical correlation) |
| Preferred regimen | Cefiderocol 2 g IV q8h (3-hour infusion) × 10–14 days (adjust per renal function) |
| Alternative regimen | If available and supported by susceptibility: ceftazidime-avibactam + aztreonam (common strategy for MBL producers) |
| Stewardship notes | Confirm susceptibility when possible; monitor renal function closely; consult ID for combination strategies |
| Source examples | IDSA guidance for difficult-to-treat Pseudomonas; FDA labeling for cefiderocol |
A simple rule format for a “predefined formulary library”
Below is a compact JSON shape you can store in a database. Your report generator can select the best matching rule(s) based on detected pathogen + AMR markers.
{
"pathogen": "Escherichia coli",
"amr_markers": ["blaCTX-M"],
"rule_scope": { "infection_site": "UTI", "setting": "inpatient|outpatient" },
"preferred_regimen": { "drug": "ertapenem", "dose": "1 g IV q24h", "duration_days": 7 },
"alternatives": [
{ "drug": "meropenem", "dose": "1 g IV q8h", "duration_days": 7 },
{
"drug": "TMP-SMX DS",
"dose": "160/800 mg PO q12h",
"duration_days": 10,
"conditions": ["if susceptible", "if clinically stable"]
}
],
"dose_adjustments": [
{ "condition": "CrCl <30 mL/min", "change": "ertapenem 500 mg IV q24h" }
],
"warnings": ["requires site/severity confirmation", "review at 48–72h for de-escalation"],
"source_notes": "IDSA resistant GN guidance; Sanford Guide (digital)"
}