Taming LLMs for Infectious-Disease Lab Reports: From Risk to Reliability
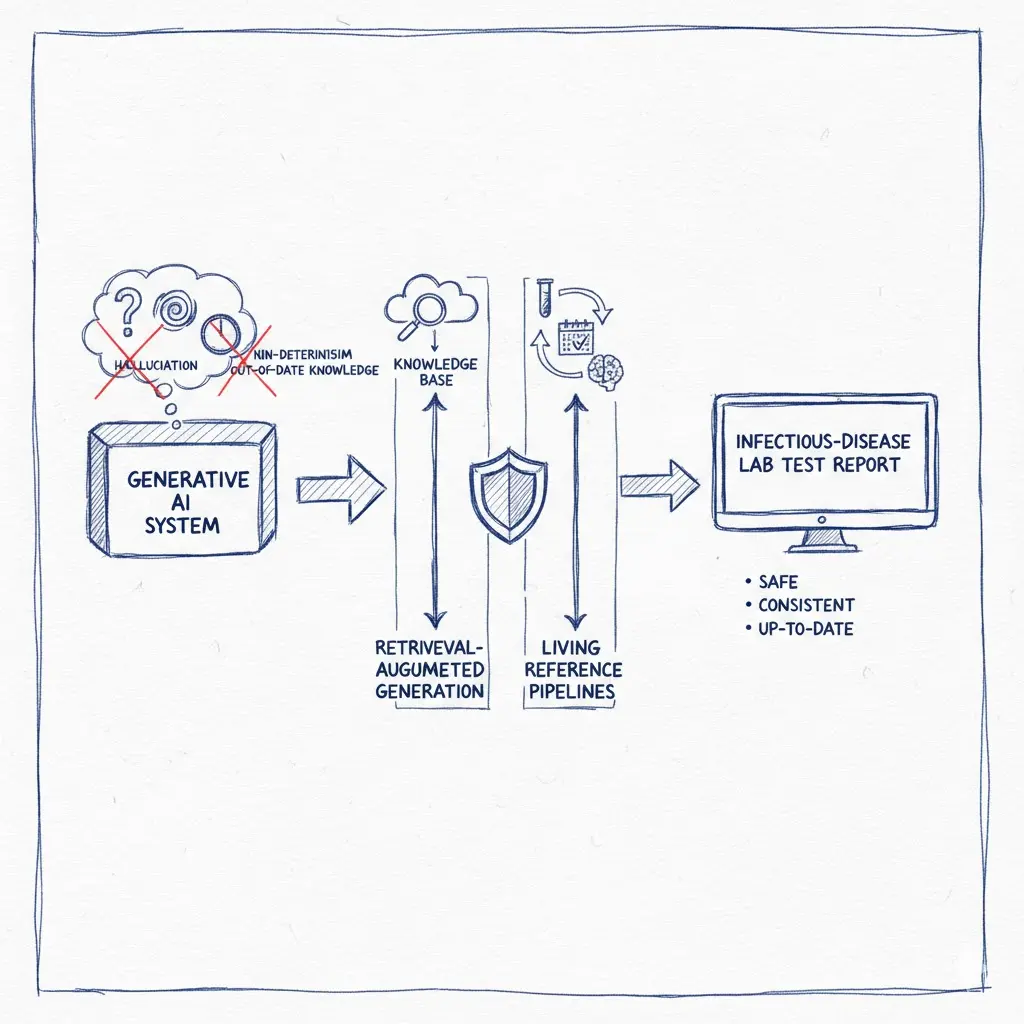
Generating infectious-disease lab test reports is a precision task. Yet out-of-the-box large language models (LLMs) can hallucinate, vary across runs, and fall behind fast-moving guidelines—three failure modes that are unacceptable in clinical settings. Below is a pragmatic blueprint to address each shortcoming with retrieval-augmented generation (RAG), deterministic engineering, and a living data pipeline anchored to authoritative sources.
1) Hallucination: Ground the model with authoritative sources (RAG)
The problem: LLMs can fabricate drug names or cite non-existent regimens. In lab reports, that’s more than a nuisance—it’s a patient-safety hazard.
The fix: Bind the model to trusted references via RAG and require provenance in every recommendation.
Build a source-of-truth corpus:
- Labeling and drug facts: Use regulatory endpoints and bulk downloads to verify drug existence, ingredients, and labeling. Pair with current package-inserts for latest data.
- Global/US treatment guidance: Include WHO’s antibiotic classification and CDC’s clinical guidance as clinical policy anchors. Where relevant, add specialist society practice guidelines.
Enforce retrieval-first generation:
- For each organism + susceptibility profile, retrieve the top K passages from the corpus; only then generate. Require the LLM to quote the specific source lines (ID, section, date).
- Add post-generation checks: confirm that any antibiotic mentioned appears in the validated set and is classified appropriately. Fail closed if not found.
2) Non-determinism: Make recommendations consistent, not creative
The problem: The same inputs can yield different outputs—fine for brainstorming, unsafe for reports.
The fix: Engineer for repeatability across retrieval, decoding, and policy application.
- Deterministic decoding: Prefer greedy or beam search (
do_sample=False) andtemperature=0to remove stochasticity. Pair with seeding and framework-level deterministic modes. - Canonical retrieval: Keep RAG inputs stable by freezing retriever settings (embedding model/version, top-K, filters, ranking). Cache “canonical context bundles” per test archetype.
- Policy via expert database: Externalize recommendations (dose, duration, alternatives, contraindications) into a versioned expert knowledge base distilled from guidance. The LLM then renders this policy table for the patient context rather than inventing regimens.
- Structured classification before generation: Use a robust classifier (e.g., BART) to map lab results to a treatment class (uncomplicated cystitis, PID, ESBL-E bacteremia, etc.), then apply the appropriate policy template.
- Response caching: Cache identical queries (and near-duplicates via semantic caching) so routine cases return the same, verified text and citations.
3) Out-of-date knowledge: Build a living pipeline that updates itself
The problem: Base LLM knowledge is always months behind—dangerous when dosing or resistance guidance changes.
The fix: Automate a data refresh → index → retrieve loop tied to authoritative endpoints.
A practical pipeline:
- Scheduled ingestion (e.g., cron/Airflow/GitHub Actions).
- Package-inserts: daily/weekly/monthly dumps of all labels.
- Regulatory APIs/data: download zipped datasets for updates.
- Clinical guidance: use content APIs to keep guidance in sync.
- Specialist society updates: monitor major guideline updates (e.g., antimicrobial-resistance treatment).
- Normalize & version. Parse to a common schema (drug → ingredient(s) → dose forms → indications → contraindications) and stamp the source date so every recommendation can be reproduced “as of” a specific day. (Provenance is your audit trail.)
- Chunk, embed, and index. Use a vector store such as FAISS, Milvus, or pgvector; choose ANN indexes (HNSW/IVF) appropriate for corpus size and latency.
- Retrieval policy. At runtime, retrieve only documents newer than the last guideline update and filter by jurisdiction (e.g., US labeling for US patients). Include the “as-of” date in the report footer.
- Human-in-the-loop & safety checks. Route novel or conflicting cases to stewardship pharmacists/ID specialists; log every source snippet and model hash for QA.
Final thoughts
For infectious-disease lab reports, accuracy, consistency, and currency are non-negotiable. Grounding with RAG, taming generation with deterministic settings and classification, and operating a living data pipeline transform LLMs from risky free-form generators into traceable clinical summarizers that cite and conform to regulatory and society sources. Always treat the model as a documentation assistant, not an autonomous prescriber, and keep a clinician in the loop for exceptions and governance.
References
- „Reducing hallucinations in structured outputs via Retrieval-Augmented Generation”, ArXiv 2024.
- „Understanding retrieval-augmented generation (RAG): a response to hallucinations”, Ontoforce Blog.
- „How to Prevent LLM Hallucinations: 5 Proven Strategies”, Voiceflow Blog.
- „Outpatient Clinical Care for Adults | Antibiotic Prescribing and Use”, CDC.
- „Infectious Diseases Society of America 2024 Guidance on the Treatment of Antimicrobial-Resistant Gram-Negative Infections”, IDSA.
- „Core Elements of Hospital Antibiotic Stewardship Programs”, CDC.
Disclaimer: This post describes engineering patterns for report generation and is not medical advice. Clinical decisions must be made by licensed professionals using full patient context.